ndsampler package¶
Subpackages¶
Submodules¶
- ndsampler.abstract_frames module
- ndsampler.abstract_sampler module
AbstractSamplerAbstractSampler.class_idsAbstractSampler.lookup_class_name()AbstractSampler.lookup_class_id()AbstractSampler.load_sample()AbstractSampler.n_positivesAbstractSampler.load_item()AbstractSampler.load_positive()AbstractSampler.load_negative()AbstractSampler.load_image()AbstractSampler.image_ids()AbstractSampler.preselect()
- ndsampler.coco_frames module
- ndsampler.coco_regions module
MissingNegativePoolTargetsCocoRegionsCocoRegions.catgraphCocoRegions.n_negativesCocoRegions.n_positivesCocoRegions.n_samplesCocoRegions.class_idsCocoRegions.image_idsCocoRegions.n_annotsCocoRegions.n_imagesCocoRegions.n_categoriesCocoRegions.lookup_class_name()CocoRegions.lookup_class_id()CocoRegions.demo()CocoRegions.isect_indexCocoRegions.targetsCocoRegions.neg_anchorsCocoRegions.overlapping_aids()CocoRegions.get_segmentations()CocoRegions.get_negative()CocoRegions.get_positive()CocoRegions.get_item()CocoRegions.new_sample_grid()
tabular_coco_targets()select_positive_regions()new_video_sample_grid()new_image_sample_grid()
- ndsampler.coco_sampler module
CocoSamplerCocoSampler.demo()CocoSampler.coerce()CocoSampler.classesCocoSampler.catgraphCocoSampler.lookup_class_name()CocoSampler.lookup_class_id()CocoSampler.n_positivesCocoSampler.n_annotsCocoSampler.n_samplesCocoSampler.n_imagesCocoSampler.n_categoriesCocoSampler.class_idsCocoSampler.image_idsCocoSampler.preselect()CocoSampler.new_sample_grid()CocoSampler.load_image_with_annots()CocoSampler.load_annotations()CocoSampler.load_image()CocoSampler.load_item()CocoSampler.load_positive()CocoSampler.load_negative()CocoSampler.load_sample()
- ndsampler.isect_indexer module
- ndsampler.toydata module
DynamicToySamplerDynamicToySampler.load_item()DynamicToySampler.class_idsDynamicToySampler.n_positivesDynamicToySampler.n_annotsDynamicToySampler.n_imagesDynamicToySampler.image_ids()DynamicToySampler.lookup_class_name()DynamicToySampler.lookup_class_id()DynamicToySampler.n_categoriesDynamicToySampler.preselect()DynamicToySampler.load_image()DynamicToySampler.load_image_with_annots()DynamicToySampler.load_sample()DynamicToySampler.load_positive()DynamicToySampler.load_negative()
Module contents¶
mkinit ~/code/ndsampler/ndsampler/__init__.py –diff mkinit ~/code/ndsampler/ndsampler/__init__.py -w
- class ndsampler.AbstractSampler[source]¶
Bases:
objectAPI for Samplers, not all methods need to be implemented depending on the use case (for example, load_sample may not be defined if positive / negative cases are generated on the fly).
- property class_ids¶
- property n_positives¶
- class ndsampler.CategoryTree(graph=None, checks=True)[source]¶
Bases:
CategoryTree,Mixin_CategoryTree_Torch- Parameters:
graph (nx.DiGraph) – either the graph representing a category hierarchy
checks (bool, default=True) – if false, bypass input checks
- class ndsampler.CocoFrames(dset, hashid_mode='PATH', workdir=None, verbose=0, backend='auto')[source]¶
Bases:
Frames,HashIdentifiablewrapper around coco-style dataset to allow for getitem syntax
CommandLine
xdoctest -m ndsampler.coco_frames CocoFrames
Example
>>> from ndsampler.coco_frames import * >>> import ndsampler >>> import kwcoco >>> import ubelt as ub >>> workdir = ub.Path.appdir('ndsampler').ensuredir() >>> dset = kwcoco.CocoDataset.demo(workdir=workdir) >>> dset._ensure_imgsize() >>> self = CocoFrames(dset, workdir=workdir) >>> assert self.load_image(1).shape == (512, 512, 3) >>> assert self.load_image(1)[:-20, :-10].shape == (492, 502, 3) >>> assert self.load_region(1, (slice(-20), slice(-10))).shape == (492, 502, 3)
Example
>>> from ndsampler import coco_sampler >>> self = coco_sampler.CocoSampler.demo().frames >>> assert self.load_image(1).shape == (600, 600, 3) >>> assert self.load_image(1)[:-20, :-10].shape == (580, 590, 3)
- property image_ids¶
- class ndsampler.CocoRegions(dset, workdir=None, verbose=1)[source]¶
Bases:
Targets,HashIdentifiable,NiceReprConverts Coco-Style datasets into a table for efficient on-line work
Perhaps rename this class to regions, and then have targets be an attribute of regions.
- Parameters:
dset (ndsampler.CocoAPI) – a dataset in coco format
workdir (PathLike) – a temporary directory where we can cache stuff
verbose (int) – verbosity level
Example
>>> from ndsampler.coco_regions import * >>> self = CocoRegions.demo() >>> pos_tr = self.get_positive(rng=0) >>> neg_tr = self.get_negative(rng=0) >>> print(ub.urepr(pos_tr, precision=2)) >>> print(ub.urepr(neg_tr, precision=2))
- property catgraph¶
- property n_negatives¶
- property n_positives¶
- property n_samples¶
- property class_ids¶
- property image_ids¶
- property n_annots¶
- property n_images¶
- property n_categories¶
- property isect_index¶
Lazy access to a disk-cached intersection index for this dataset
- property targets¶
All viable positive annotations targets in a flat table.
The main idea is that this is the population of all positives that we could sample from. Often times we will simply use all of them.
This function takes a subset of annotations in the coco dataset that can be considered “viable” positives. We may subsample these futher, but this serves to collect the annotations that could feasibly be used by the network. Essentailly we remove annotations without bounding boxes. I’m not sure I 100% like the way this works though. Shouldn’t filtering be done before we even get here? Perhaps but perhaps not. This design needs a bit more thought.
- property neg_anchors¶
- overlapping_aids(gid, region, visible_thresh=0.0)[source]¶
Finds the other annotations in this image that overlap a region
- Parameters:
gid (int) – image id
region (kwimage.Boxes) – bounding box
visible_thresh (float) – does not return annotations with visibility less than this threshold.
- Returns:
annotation ids
- Return type:
List[int]
- get_segmentations(aids)[source]¶
Returns the segmentations corresponding to a set of annotation ids
Example
>>> from ndsampler.coco_regions import * >>> from ndsampler import coco_sampler >>> self = coco_sampler.CocoSampler.demo().regions >>> aids = [1, 2]
- get_negative(index=None, rng=None)[source]¶
Get localization information for a negative region
- Parameters:
index (int or None) – indexes into the current negative pool or if None returns a random negative
rng (RandomState) – used only if index is None
- Returns:
tr: target info dictionary
- Return type:
Dict
CommandLine
xdoctest -m ndsampler.coco_regions CocoRegions.get_negative
Example
>>> from ndsampler.coco_regions import * >>> from ndsampler import coco_sampler >>> rng = kwarray.ensure_rng(0) >>> self = coco_sampler.CocoSampler.demo().regions >>> tr = self.get_negative(rng=rng) >>> # xdoctest: +IGNORE_WANT >>> assert 'category_id' in tr >>> assert 'aid' in tr >>> assert 'cx' in tr >>> print(ub.urepr(tr, precision=2)) { 'aid': -1, 'category_id': 0, 'cx': 190.71, 'cy': 95.83, 'gid': 1, 'height': 140.00, 'img_height': 600, 'img_width': 600, 'width': 68.00, }
- get_positive(index=None, rng=None)[source]¶
Get localization information for a positive region
- Parameters:
index (int or None) – indexes into the current positive pool or if None returns a random negative
rng (RandomState) – used only if index is None
- Returns:
tr: target info dictionary
- Return type:
Dict
Example
>>> from ndsampler import coco_sampler >>> rng = kwarray.ensure_rng(0) >>> self = coco_sampler.CocoSampler.demo().regions >>> tr = self.get_positive(0, rng=rng) >>> print(ub.urepr(tr, precision=2))
- new_sample_grid(task, window_dims, window_overlap=0, **kwargs)[source]¶
New experimental method to replace preselect positives / negatives
- Parameters:
task (str) – can be video_detection image_detection # video_classification # image_classification
**kwargs – passed to new_video_sample_grid or new_image_sample_grid
Example
>>> from ndsampler.coco_regions import * >>> from ndsampler import coco_sampler >>> self = coco_sampler.CocoSampler.demo('vidshapes1').regions >>> self.dset.conform() >>> sample_grid = self.new_sample_grid('video_detection', window_dims=(2, 100, 100))
- class ndsampler.CocoSampler(dset, workdir=None, autoinit=True, backend=None, verbose=0)[source]¶
Bases:
AbstractSampler,HashIdentifiable,NiceReprSamples patches of positives and negative detection windows from a COCO dataset. Can be used for training FCN or RPN based classifiers / detectors.
Does data loading, padding, etc…
- Parameters:
dset (kwcoco.CocoDataset) – a coco-formatted dataset
backend (str | Dict) – Can be None, ‘cog’ or ‘npy’, or a dict. In the case of a dict, it takes the format: {‘type’: str, ‘config’: Dict}. See AbstractFrames for more details. Defaults to None, which does not do anything fancy.
Example
#print >>> from ndsampler.coco_sampler import * >>> self = CocoSampler.demo(‘photos’) … >>> print(sorted(self.class_ids)) [0, 1, 2, 3, 4, 5, 6, 7, 8] >>> print(self.n_positives) 4
Example
>>> import ndsampler >>> self = ndsampler.CocoSampler.demo('photos') >>> p_sample = self.load_positive() >>> n_sample = self.load_negative() >>> self = ndsampler.CocoSampler.demo('shapes') >>> p_sample2 = self.load_positive() >>> n_sample2 = self.load_negative() >>> for sample in [p_sample, n_sample, p_sample2, n_sample2]: >>> assert 'annots' in sample >>> assert 'im' in sample >>> assert 'rel_boxes' in sample['annots'] >>> assert 'rel_ssegs' in sample['annots'] >>> assert 'rel_kpts' in sample['annots'] >>> assert 'cids' in sample['annots'] >>> assert 'aids' in sample['annots']
- classmethod demo(key='shapes', workdir=None, backend=None, **kw)[source]¶
Create a toy coco sampler for testing and demo puposes
- SeeAlso:
kwcoco.CocoDataset.demo
- classmethod coerce(data, **kwargs)[source]¶
Attempt to coerce the input data into a sampler. Generally this can be anything that is already a sampler, or somthing that can be coerced into a kwcoco dataset.
- Parameters:
data (str | PathLike | CocoDataset | CocoSampler) – something that can be coerced into a CocoSampler.
- Returns:
CocoSampler
- property classes¶
- property catgraph¶
DEPRICATED, use self.classes instead
- property n_positives¶
- property n_annots¶
- property n_samples¶
- property n_images¶
- property n_categories¶
- property class_ids¶
- property image_ids¶
- load_image_with_annots(image_id, cache=True)[source]¶
- Parameters:
image_id (int) – the coco image id
cache (bool) – if True returns the fast subregion-indexable file reference. Otherwise, eagerly loads the entire image. Defaults to True.
- Returns:
img: the coco image dict augmented with imdata anns: the coco annotations in this image
- Return type:
Tuple[Dict, List[Dict]]
Example
>>> import ndsampler >>> self = ndsampler.CocoSampler.demo() >>> img, anns = self.load_image_with_annots(1) >>> dets = kwimage.Detections.from_coco_annots(anns, dset=self.dset) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> kwplot.imshow(img['imdata'][:], doclf=1) >>> dets.draw() >>> kwplot.show_if_requested()
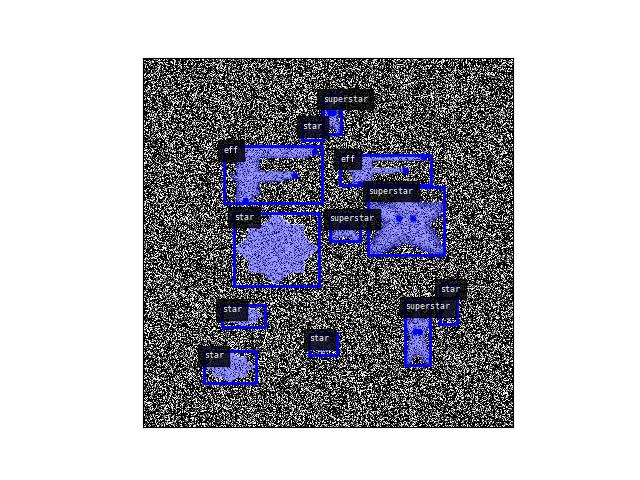
- load_annotations(image_id)[source]¶
Loads the annotations within an image
- Parameters:
image_id (int) – the coco image id
- Returns:
list of coco annotation dictionaries
- Return type:
List[Dict]
- load_image(image_id, cache=True)[source]¶
Loads the annotations within an image
- Parameters:
image_id (int) – the coco image id
cache (bool) – if True returns the fast subregion-indexable file reference. Otherwise, eagerly loads the entire image. Defaults to True.
- Returns:
either ndarray data or a indexable reference
- Return type:
ArrayLike
- load_item(index, with_annots=True, target=None, rng=None, **kw)[source]¶
Loads item from either positive or negative regions pool.
Lower indexes will return positive regions and higher indexes will return negative regions.
The main paradigm of the sampler is that sampler.regions maintains a pool of target regions, you can influence what that pool is at any point by calling sampler.regions.preselect (usually either at the start of learning, or maybe after every epoch, etc..), and you use load_item to load the index-th item from that preselected pool. Depending on how you preselected the pool, the returned item might correspond to a positive or negative region.
- Parameters:
index (int) – index of target region
with_annots (bool | str) – if True, also extracts information about any annotation that overlaps the region of interest (subject to visibility_thresh). Can also be a List[str] that specifies which specific subinfo should be extracted. Valid strings in this list are: boxes, keypoints, and segmenation. Defaults to True.
target (Dict) – Extra target arguments that update the positive target, like window_dims, pad, etc…. See
load_sample()for details on allowed keywords.rng (None | int | RandomState) – a seed or seeded random number generator.
**kw – other arguments that can be passed to
CocoSampler.load_sample()
- Returns:
- sample: dict containing keys
im (ndarray): image data target (dict): contains the same input items as the input
target but additionally specifies inferred information like rel_cx and rel_cy, which gives the center of the target w.r.t the returned padded sample.
annots (dict): Dict of aids, cids, and rel/abs boxes
- Return type:
Dict
- load_positive(index=None, with_annots=True, target=None, rng=None, **kw)[source]¶
Load an item from the the positive pool of regions.
- Parameters:
index (int) – index of positive target
with_annots (bool | str) – if True, also extracts information about any annotation that overlaps the region of interest (subject to visibility_thresh). Can also be a List[str] that specifies which specific subinfo should be extracted. Valid strings in this list are: boxes, keypoints, and segmentation. Defaults to True.
target (Dict) – Extra target arguments that update the positive target, like window_dims, pad, etc…. See
load_sample()for details on allowed keywords.rng (None | int | RandomState) – a seed or seeded random number generator.
**kw – other arguments that can be passed to
CocoSampler.load_sample()
- Returns:
- sample: dict containing keys
im (ndarray): image data tr (dict): contains the same input items as tr but additionally
specifies rel_cx and rel_cy, which gives the center of the target w.r.t the returned padded sample.
annots (dict): Dict of aids, cids, and rel/abs boxes
- Return type:
Dict
Example
>>> import ndsampler >>> self = ndsampler.CocoSampler.demo() >>> sample = self.load_positive(pad=(10, 10), tr=dict(window_dims=(3, 3))) >>> assert sample['im'].shape[0] == 23 >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> kwplot.imshow(sample['im'], doclf=1) >>> kwplot.show_if_requested()
- load_negative(index=None, with_annots=True, target=None, rng=None, **kw)[source]¶
Load an item from the the negative pool of regions.
- Parameters:
index (int) – if specified loads a specific negative from the presampled pool, otherwise the next negative in the pool is returned.
with_annots (bool | str) – if True, also extracts information about any annotation that overlaps the region of interest (subject to visibility_thresh). Can also be a List[str] that specifies which specific subinfo should be extracted. Valid strings in this list are: boxes, keypoints, and segmentation. Defaults to True.
target (Dict) – Extra target arguments that update the positive target, like window_dims, pad, etc…. See
load_sample()for details on allowed keywords.rng (None | int | RandomState) – a seed or seeded random number generator.
- Returns:
- sample: dict containing keys
im (ndarray): image data tr (dict): contains the same input items as tr but additionally
specifies rel_cx and rel_cy, which gives the center of the target w.r.t the returned padded sample.
annots (dict): Dict of aids, cids, and rel/abs boxes
- Return type:
Dict
Example
>>> import ndsampler >>> self = ndsampler.CocoSampler.demo() >>> rng = None >>> sample = self.load_negative(rng=rng, pad=(0, 0)) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> import kwimage >>> kwplot.autompl() >>> abs_sample_box = sample['params']['sample_tlbr'] >>> tf_rel_from_abs = kwimage.Affine.coerce(sample['params']['tf_rel_to_abs']).inv() >>> wh, ww = sample['target']['window_dims'] >>> abs_window_box = kwimage.Boxes([[sample['target']['cx'], sample['target']['cy'], ww, wh]], 'cxywh') >>> rel_window_box = abs_window_box.warp(tf_rel_from_abs) >>> rel_sample_box = abs_sample_box.warp(tf_rel_from_abs) >>> kwplot.imshow(sample['im'], fnum=1, doclf=True) >>> rel_sample_box.draw(color='kw_green', lw=10) >>> rel_window_box.draw(color='kw_blue', lw=8) >>> kwplot.show_if_requested()
Example
>>> import ndsampler >>> self = ndsampler.CocoSampler.demo() >>> rng = None >>> sample = self.load_negative(rng=rng, pad=(10, 20), target=dict(window_dims=(64, 64))) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> import kwimage >>> kwplot.autompl() >>> abs_sample_box = sample['params']['sample_tlbr'] >>> tf_rel_from_abs = kwimage.Affine.coerce(sample['params']['tf_rel_to_abs']).inv() >>> wh, ww = sample['target']['window_dims'] >>> abs_window_box = kwimage.Boxes([[sample['target']['cx'], sample['target']['cy'], ww, wh]], 'cxywh') >>> rel_window_box = abs_window_box.warp(tf_rel_from_abs) >>> rel_sample_box = abs_sample_box.warp(tf_rel_from_abs) >>> kwplot.imshow(sample['im'], fnum=1, doclf=True) >>> rel_sample_box.draw(color='kw_green', lw=10) >>> rel_window_box.draw(color='kw_blue', lw=8) >>> kwplot.show_if_requested()
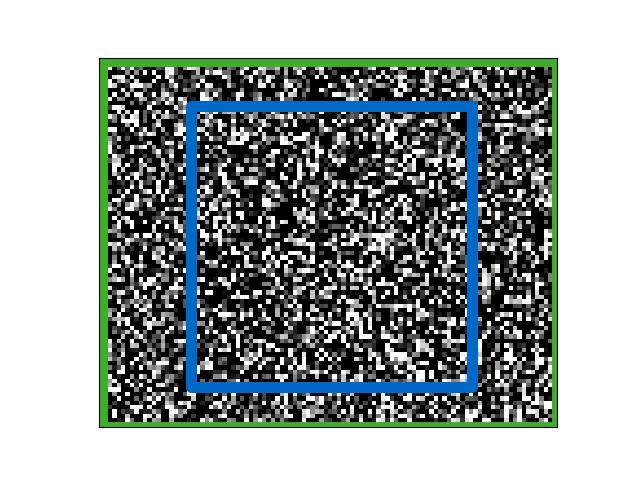
- load_sample(target=None, with_annots=True, annot_ids=None, visible_thresh=0.0, **kwargs)[source]¶
Loads the volume data associated with the bbox and frame of a target
- Parameters:
target (dict) – target dictionary (often abbreviated as tr) indicating an nd source object (e.g. image or video) and the coordinate region to sample from. Unspecified coordinate regions default to the extent of the source object.
For 2D image source objects, target must contain or be able to infer the key gid (int), to specify an image id.
For 3D video source objects, target must contain the key vidid (int), to specify a video id (NEW in 0.6.1) or gids List[int], as a list of images in a video (NEW in 0.6.2)
In general, coordinate regions can specified by the key slices, a numpy-like “fancy index” over each of the n dimensions. Usually this is a tuple of slices, e.g. (y1:y2, x1:x2) for images and (t1:t2, y1:y2, x1:x2) for videos.
You may also specify: space_slice as (y1:y2, x1:x2) for both 2D images and 3D videos and time_slice as t1:t2 for 3D videos.
- Spatial regions can be specified with keys:
‘cx’ and ‘cy’ as the center of the region in pixels.
‘width’ and ‘height’ are in pixels.
‘window_dims’ is a height, width tuple or can be a
special string key ‘square’, which overrides width and height to both be the maximum of the two.
Temporal regions are specifiable by slices, time_slice or an explicit list of gids.
The aid key can be specified to indicate a specific annotation to load. This uses the annotation information to infer ‘gid’, ‘cx’, ‘cy’, ‘width’, and ‘height’ if they are not present. (NEW in 0.5.10)
- The channels key can be specified as a channel code or
kwcoco.ChannelSpecobject. (NEW in 0.6.1)- as_xarray (bool):
if True, return the image data as an xarray object. default=False
- interpolation (str):
type of resample interpolation. Defaults to ‘auto’.
- antialias (str):
antialias sample or not. Defaults to ‘auto’.
nodata: override function level nodata
- use_native_scale (bool):
If True, the “im” field is returned as a jagged list of data that are as close to native resolution as possible while still maintaining alignment up to a scale factor. Currently only available for video sampling.
- scale (float | Tuple[float, float]):
if specified, the same window is sampled, but the data is returned warped by the extra scale factor. This augments the existing image or video scale factor. Any annotations are also warped according to this factor such that they align with the returned data. By default this scale is applied to videospace, unless use_native_scale is given, in which case it is applied to the native resolution (generally you dont want to combine these).
- pad (tuple): (height, width) extra context to add to window dims.
This helps prevent augmentation from producing boundary effects
- padkw (dict): kwargs for numpy.pad.
Defaults to {‘mode’: ‘constant’}.
- dtype (type | None):
Cast the loaded data to this type. If unspecified returns the data as-is.
- nodata (int | None):
If specified, for integer data with nodata values, this is passed to kwcoco delayed image finalize. The data is converted to float32 and nodata values are replaced with nan. These nan values are handled correctly in subsequent warping operations. Defaults to None.
with_annots (bool | str) – if True, also extracts information about any annotation that overlaps the region of interest (subject to visibility_thresh). Can also be a List[str] that specifies which specific subinfo should be extracted. Valid strings in this list are: boxes, keypoints, and segmentation. Defaults to True.
annot_ids (List[int]) – if specified, assume the user has precomputed which annotations should be loaded for the target region. Skip the spatial lookup step and just load the data for these annotations instead.
visible_thresh (float) – does not return annotations with visibility less than this threshold.
**kwargs – handles deprecated arguments which are now specified in the target dictionary itself.
- Returns:
- sample: dict containing keys
im (ndarray | DataArray): image / video data target (dict): contains the same input items as the input
target but additionally specifies inferred information like rel_cx and rel_cy, which gives the center of the target w.r.t the returned padded sample.
- annots (dict): containing items:
- frame_dets (List[kwimage.Detections]): a list of detection
objects containing the requested annotation info for each frame.
aids (list): annotation ids DEPRECATED cids (list): category ids DEPRECATED rel_ssegs (ndarray): segmentations relative to the sample DEPRECATED rel_kpts (ndarray): keypoints relative to the sample DEPRECATED
- Return type:
Dict
CommandLine
xdoctest -m ndsampler.coco_sampler CocoSampler.load_sample:2 --show xdoctest -m ndsampler.coco_sampler CocoSampler.load_sample:1 --show xdoctest -m ndsampler.coco_sampler CocoSampler.load_sample:3 --show
Example
>>> import ndsampler >>> self = ndsampler.CocoSampler.demo() >>> # The target (target) lets you specify an arbitrary window >>> target = {'gid': 1, 'cx': 5, 'cy': 2, 'width': 6, 'height': 6} >>> sample = self.load_sample(target) ... >>> print('sample.shape = {!r}'.format(sample['im'].shape)) sample.shape = (6, 6, 3)
Example
>>> # Access direct annotation information >>> import ndsampler >>> sampler = ndsampler.CocoSampler.demo() >>> # Sample a region that contains at least one annotation >>> target = {'gid': 1, 'cx': 5, 'cy': 2, 'width': 600, 'height': 600} >>> sample = sampler.load_sample(target) >>> annotation_ids = sample['annots']['aids'] >>> aid = annotation_ids[0] >>> # Method1: Access ann dict directly via the coco index >>> ann = sampler.dset.anns[aid] >>> # Method2: Access ann objects via annots method >>> dets = sampler.dset.annots(annotation_ids).detections >>> print('dets.data = {}'.format(ub.urepr(dets.data, nl=1)))
Example
>>> import ndsampler >>> self = ndsampler.CocoSampler.demo() >>> target = self.regions.get_positive(0) >>> target['window_dims'] = 'square' >>> target['pad'] = (25, 25) >>> sample = self.load_sample(target) >>> print('im.shape = {!r}'.format(sample['im'].shape)) im.shape = (135, 135, 3) >>> target['window_dims'] = None >>> target['pad'] = (0, 0) >>> sample = self.load_sample(target) >>> print('im.shape = {!r}'.format(sample['im'].shape)) im.shape = (52, 85, 3) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> kwplot.imshow(sample['im']) >>> kwplot.show_if_requested()

Example
>>> # sample an out of bounds target >>> from ndsampler.coco_sampler import * >>> self = CocoSampler.demo('vidshapes8') >>> test_vidspace = 1 >>> target = self.regions.get_positive(0) >>> # Toggle to see if this test works in both cases >>> space = 'image' >>> if test_vidspace: >>> space = 'video' >>> target = target.copy() >>> target['gids'] = [target.pop('gid')] >>> target['scale'] = 1.3 >>> #target['scale'] = 0.8 >>> #target['use_native_scale'] = True >>> #target['realign_native'] = 'largest' >>> target['window_dims'] = (364, 364) >>> sample = self.load_sample(target) >>> annots = sample['annots'] >>> assert len(annots['aids']) > 0 >>> #assert len(annots['rel_cxywh']) == len(annots['aids']) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> tf_rel_to_abs = sample['params']['tf_rel_to_abs'] >>> rel_dets = annots['frame_dets'][0] >>> abs_dets = rel_dets.warp(tf_rel_to_abs) >>> # Draw box in original image context >>> #abs_frame = self.frames.load_image(sample['target']['gid'], space=space)[:] >>> abs_frame = self.dset.coco_image(sample['target']['gid']).delay(space=space).finalize() >>> kwplot.imshow(abs_frame, pnum=(1, 2, 1), fnum=1) >>> abs_dets.data['boxes'].translate([-.5, -.5]).draw() >>> abs_dets.data['keypoints'].draw(color='green', radius=10) >>> abs_dets.data['segmentations'].draw(color='red', alpha=.5) >>> # Draw box in relative sample context >>> if test_vidspace: >>> kwplot.imshow(sample['im'][0], pnum=(1, 2, 2), fnum=1) >>> else: >>> kwplot.imshow(sample['im'], pnum=(1, 2, 2), fnum=1) >>> rel_dets.data['boxes'].translate([-.5, -.5]).draw() >>> rel_dets.data['segmentations'].draw(color='red', alpha=.6) >>> rel_dets.data['keypoints'].draw(color='green', alpha=.4, radius=10) >>> kwplot.show_if_requested()
Example
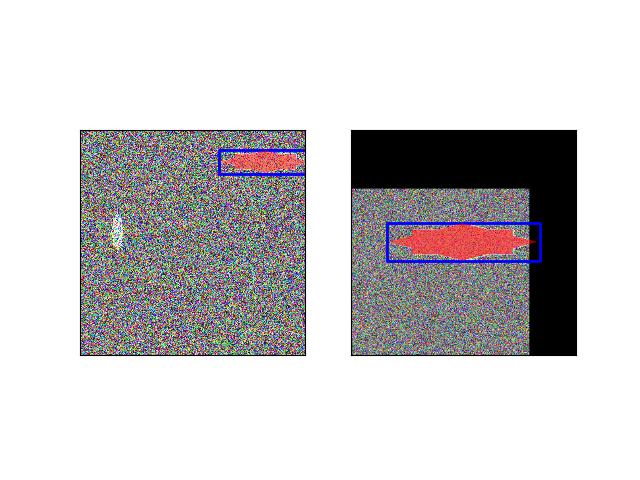
>>> from ndsampler.coco_sampler import * >>> self = CocoSampler.demo('photos') >>> target = self.regions.get_positive(1) >>> target['window_dims'] = (300, 150) >>> target['pad'] = None >>> sample = self.load_sample(target) >>> assert sample['im'].shape[0:2] == target['window_dims'] >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> kwplot.imshow(sample['im'], colorspace='rgb') >>> kwplot.show_if_requested()

Example
>>> # Multispectral video sample example >>> from ndsampler.coco_sampler import * >>> self = CocoSampler.demo('vidshapes1-multispectral', num_frames=5) >>> sample_grid = self.new_sample_grid('video_detection', (3, 128, 128)) >>> target = sample_grid['positives'][0] >>> target['channels'] = 'B1|B8' >>> target['as_xarray'] = False >>> sample = self.load_sample(target) >>> print(ub.urepr(sample['target'], nl=1)) >>> print(sample['im'].shape) >>> assert sample['im'].shape == (3, 128, 128, 2) >>> target['channels'] = '<all>' >>> sample = self.load_sample(target) >>> assert sample['im'].shape == (3, 128, 128, 5)
Example
>>> # Multispectral-multisensor jagged video sample example >>> from ndsampler.coco_sampler import * >>> self = CocoSampler.demo('vidshapes1-msi-multisensor', num_frames=5) >>> sample_grid = self.new_sample_grid('video_detection', (3, 128, 128)) >>> target = sample_grid['positives'][0] >>> target['channels'] = 'B1|B8' >>> target['as_xarray'] = False >>> sample1 = self.load_sample(target) >>> target['scale'] = 2 >>> sample2 = self.load_sample(target) >>> target['use_native_scale'] = True >>> sample3 = self.load_sample(target) >>> #### >>> assert sample1['im'].shape == (3, 128, 128, 2) >>> assert sample2['im'].shape == (3, 256, 256, 2) >>> box1 = sample1['annots']['frame_dets'][0].boxes >>> box2 = sample2['annots']['frame_dets'][0].boxes >>> box3 = sample3['annots']['frame_dets'][0].boxes >>> assert np.allclose((box2.width / box1.width), 2) >>> # Jagged annotations are still in video space >>> assert np.allclose((box3.width / box1.width), 2) >>> jagged_shape = [[p.shape for p in f] for f in sample3['im']] >>> jagged_align = [[a for a in m['align']] for m in sample3['params']['jagged_meta']]
- class ndsampler.DynamicToySampler(n_positives=100000.0, seed=None, gsize=(416, 416), categories=None)[source]¶
Bases:
AbstractSamplerGenerates positive and negative samples on the fly.
Note
Its probably more robust to generate a static fixed-size dataset with ‘demodata_toy_dset’ or kwcoco.CocoDataset.demo. However, if you need a sampler that dynamically generates toydata, this is for you.
CommandLine
xdoctest -m ndsampler.toydata DynamicToySampler --show
Example
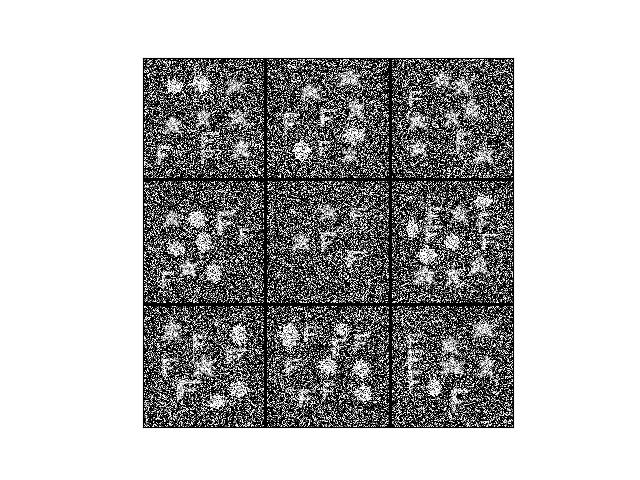
>>> # Test that this sampler works with the dataset >>> from ndsampler.toydata import * >>> self = DynamicToySampler(1e3) >>> imgs = [self.load_positive()['im'] for _ in range(9)] >>> # xdoctest: +REQUIRES(--show) >>> stacked = kwimage.stack_images_grid(imgs, overlap=-10) >>> import kwplot >>> kwplot.autompl() >>> kwplot.imshow(stacked) >>> kwplot.show_if_requested()
- property class_ids¶
- property n_positives¶
- property n_annots¶
- property n_images¶
- property n_categories¶
- preselect(n_pos=None, n_neg=None, neg_to_pos_ratio=None, window_dims=None, rng=None, verbose=0)[source]¶
- load_positive(index=None, pad=None, window_dims=None, rng=None)[source]¶
Note: window_dims is height / width
Example
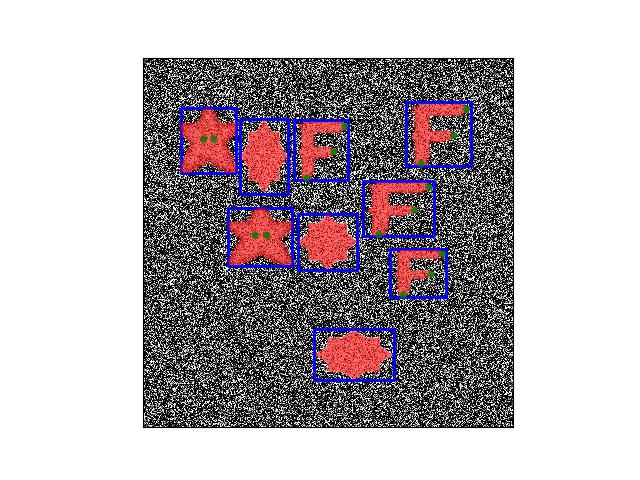
>>> from ndsampler.toydata import * >>> self = DynamicToySampler(1e2) >>> sample = self.load_positive() >>> annots = sample['annots'] >>> assert len(annots['aids']) > 0 >>> assert len(annots['rel_cxywh']) == len(annots['aids']) >>> # xdoc: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> # Draw box in relative sample context >>> kwplot.imshow(sample['im'], pnum=(1, 1, 1), fnum=1) >>> annots['rel_boxes'].translate([-.5, -.5]).draw() >>> annots['rel_ssegs'].draw(color='red', alpha=.6) >>> annots['rel_kpts'].draw(color='green', alpha=.8, radius=4)
- class ndsampler.FrameIntersectionIndex[source]¶
Bases:
NiceReprBuild spatial tree for each frame so we can quickly determine if a random negative is too close to a positive. For each frame/image we built a qtree.
Example
>>> from ndsampler.isect_indexer import * >>> import kwcoco >>> import ubelt as ub >>> dset = kwcoco.CocoDataset.demo() >>> dset._ensure_imgsize() >>> dset.remove_annotations([ann for ann in dset.anns.values() >>> if 'bbox' not in ann]) >>> # Build intersection index aroung coco dataset >>> self = FrameIntersectionIndex.from_coco(dset) >>> gid = 1 >>> box = kwimage.Boxes([0, 10, 100, 100], 'xywh') >>> isect_aids, ious = self.ious(gid, box) >>> print(ub.urepr(ious.tolist(), nl=0, precision=4)) [0.0507]
- classmethod from_coco(dset, verbose=0)[source]¶
- Parameters:
dset (kwcoco.CocoDataset) – positive annotation data
- Returns:
FrameIntersectionIndex
- classmethod demo(*args, **kwargs)[source]¶
Create a demo intersection index.
- Parameters:
*args – see kwcoco.CocoDataset.demo
**kwargs – see kwcoco.CocoDataset.demo
- Returns:
FrameIntersectionIndex
- overlapping_aids(gid, box)[source]¶
Find all annotation-ids within an image that have some overlap with a bounding box.
- Parameters:
gid (int) – an image id
box (kwimage.Boxes) – the specified region
- Returns:
list of annotation ids
- Return type:
List[int]
CommandLine
USE_RTREE=0 xdoctest -m ndsampler.isect_indexer FrameIntersectionIndex.overlapping_aids USE_RTREE=1 xdoctest -m ndsampler.isect_indexer FrameIntersectionIndex.overlapping_aids
Example
>>> from ndsampler.isect_indexer import * # NOQA >>> self = FrameIntersectionIndex.demo('shapes128') >>> for gid, qtree in self.qtrees.items(): >>> box = kwimage.Boxes([0, 0, qtree.width, qtree.height], 'xywh') >>> print(self.overlapping_aids(gid, box))
- ious(gid, box)[source]¶
Find overlaping annotations in a specific image and their intersection over union with a a query box.
- Parameters:
gid (int) – an image id
box (kwimage.Boxes) – the specified region
- Returns:
isect_aids: list of annotation ids ious: jaccard score for each returned annotation id
- Return type:
Tuple[List[int], ndarray]
- iooas(gid, box)[source]¶
Intersection over other’s area
- Parameters:
gid (int) – an image id
box (kwimage.Boxes) – the specified region
Like iou, but non-symetric, returned number is a percentage of the other’s (groundtruth) area. This means we dont care how big the (negative) box is.
- random_negatives(num, anchors=None, window_size=None, gids=None, thresh=0.0, exact=True, rng=None, patience=None)[source]¶
Finds random boxes that don’t have a large overlap with positive instances.
- Parameters:
num (int) – number of negative boxes to generate (actual number of boxes returned may be less unless exact=True)
anchors (ndarray) – prior normalized aspect ratios for negative boxes. Mutually exclusive with window_size.
window_size (ndarray) – absolute (W, H) sizes to use for negative boxes. Mutually exclusive with anchors.
gids (List[int]) – image-ids to generate negatives for, if not specified generates for all images.
thresh (float) – overlap area threshold as a percentage of the negative box size. When thresh=0.0, that means negatives cannot overlap any positive, when threh=1.0, there are no constrains on negative placement.
exact (bool) – if True, ensure that we generate exactly num boxes
rng (RandomState) – random number generator
Example
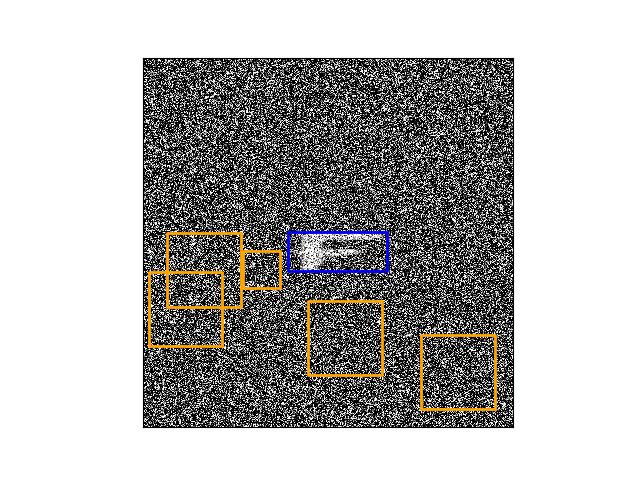
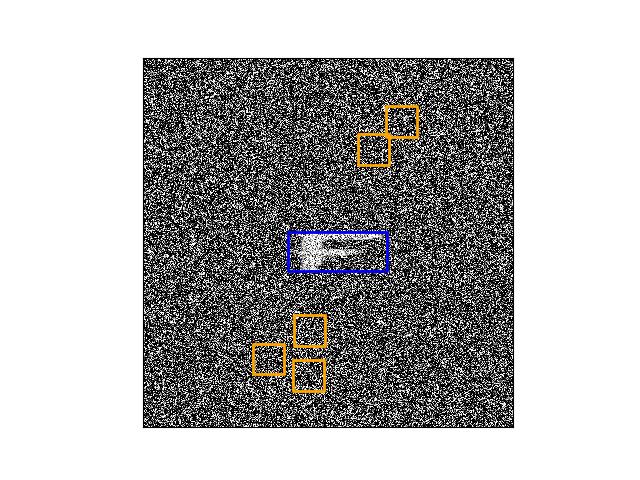
>>> from ndsampler.isect_indexer import * >>> import ndsampler >>> import kwcoco >>> dset = kwcoco.CocoDataset.demo('shapes8') >>> self = FrameIntersectionIndex.from_coco(dset) >>> anchors = np.array([[.35, .15], [.2, .2], [.1, .1]]) >>> #num = 25 >>> num = 5 >>> rng = kwarray.ensure_rng(None) >>> neg_gids, neg_boxes = self.random_negatives( >>> num, anchors, gids=[1], rng=rng, thresh=0.01, exact=1) >>> # xdoc: +REQUIRES(--show) >>> gid = sorted(set(neg_gids))[0] >>> boxes = neg_boxes.compress(neg_gids == gid) >>> import kwplot >>> kwplot.autompl() >>> img = kwimage.imread(dset.imgs[gid]['file_name']) >>> kwplot.imshow(img, doclf=True, fnum=1, colorspace='bgr') >>> support = self._support(gid) >>> kwplot.draw_boxes(support, color='blue') >>> kwplot.draw_boxes(boxes, color='orange')
Example
>>> from ndsampler.isect_indexer import * >>> import kwcoco >>> dset = kwcoco.CocoDataset.demo('shapes8') >>> self = FrameIntersectionIndex.from_coco(dset) >>> #num = 25 >>> num = 5 >>> rng = kwarray.ensure_rng(None) >>> window_size = (50, 50) >>> neg_gids, neg_boxes = self.random_negatives( >>> num, window_size=window_size, gids=[1], rng=rng, >>> thresh=0.01, exact=1) >>> # xdoc: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> gid = sorted(set(neg_gids))[0] >>> boxes = neg_boxes.compress(neg_gids == gid) >>> img = kwimage.imread(dset.imgs[gid]['file_name']) >>> kwplot.imshow(img, doclf=True, fnum=1, colorspace='bgr') >>> support = self._support(gid) >>> support.draw(color='blue') >>> boxes.draw(color='orange')
- class ndsampler.Frames(hashid_mode='PATH', workdir=None, backend=None)[source]¶
Bases:
objectAbstract implementation of Frames.
While this is an abstract class, it contains most of the
Framesfunctionality. The inheriting class needs to overload the constructor and_lookup_gpath, which maps an image-id to its path on disk.- Parameters:
hashid_mode (str, default=’PATH’) – The method used to compute a unique identifier for every image. to can be PATH, PIXELS, or GIVEN. TODO: Add DVC as a method (where it uses the name of the symlink)?
workdir (PathLike) – This is the directory where Frames can store cached results. This SHOULD be specified.
backend (str | Dict) – Determine the backend to use for fast subimage region lookups. This can either be a string ‘cog’ or ‘npy’. This can also be a config dictionary for fine-grained backend control. For this case, ‘type’: specified cog or npy, and only COG has additional options which are:
- {
‘type’: ‘cog’, ‘config’: { ‘compress’: <’LZW’ | ‘JPEG | ‘DEFLATE’ | ‘ZSTD’ | ‘auto’>, }
}
Example
>>> from ndsampler.abstract_frames import * >>> self = SimpleFrames.demo(backend='npy') >>> file = self.load_image(1) >>> print('file = {!r}'.format(file)) >>> assert self.load_image(1).shape == (512, 512, 3) >>> assert self.load_region(1, (slice(-20), slice(-10))).shape == (492, 502, 3) >>> # xdoctest: +REQUIRES(module:osgeo) >>> self = SimpleFrames.demo(backend='cog') >>> assert self.load_image(1).shape == (512, 512, 3) >>> assert self.load_region(1, (slice(-20), slice(-10))).shape == (492, 502, 3)
Benchmark
>>> from ndsampler.abstract_frames import * # NOQA >>> import ubelt as ub >>> # >>> ti = ub.Timerit(100, bestof=3, verbose=2) >>> # >>> self = SimpleFrames.demo(backend='cog') >>> for timer in ti.reset('cog-small-subregion'): >>> self.load_image(1)[10:42, 10:42] >>> # >>> self = SimpleFrames.demo(backend='npy') >>> for timer in ti.reset('npy-small-subregion'): >>> self.load_image(1)[10:42, 10:42] >>> print('----') >>> # >>> self = SimpleFrames.demo(backend='cog') >>> for timer in ti.reset('cog-large-subregion'): >>> self.load_image(1)[3:-3, 3:-3] >>> # >>> self = SimpleFrames.demo(backend='npy') >>> for timer in ti.reset('npy-large-subregion'): >>> self.load_image(1)[3:-3, 3:-3] >>> print('----') >>> # >>> self = SimpleFrames.demo(backend='cog') >>> for timer in ti.reset('cog-loadimage'): >>> self.load_image(1) >>> # >>> self = SimpleFrames.demo(backend='npy') >>> for timer in ti.reset('npy-loadimage'): >>> self.load_image(1)
- DEFAULT_NPY_CONFIG = {'config': {}, 'type': 'npy'}¶
- DEFAULT_COG_CONFIG = {'_hack_use_cli': True, 'config': {'compress': 'auto'}, 'type': 'cog'}¶
- property cache_dpath¶
Returns the path where cached frame representations will be stored.
This will be None if there is no backend.
- property image_ids¶
- load_region(image_id, region=None, channels=NoParam, width=None, height=None)[source]¶
Ammortized O(1) image subregion loading (assuming constant region size)
if region size is varied, then sampling time scales with the number of tiles needed to overlap the requested region.
- Parameters:
image_id (int) – image identifier
region (Tuple[slice, …]) – space-time region within an image
channels (str) – NotImplemented
width (int) – if the width of the entire image is know specify it
height (int) – if the height of the entire image is know specify it
- load_image(image_id, channels=NoParam, cache=True, noreturn=False)[source]¶
Load the image data for a particular image id
- Parameters:
image_id (int) – the id of the image to load
cache (bool, default=True) – ensure and return the efficient backend cached representation.
channels – NotImplemented
noreturn (bool, default=False) – if True, nothing is returned. This is useful if you simply want to ensure the cached representation.
CAREFUL: THIS NEEDS TO MAINTAIN A STABLE API. OTHER PROJECTS DEPEND ON IT.
- Returns:
- an indexable array like representation, possibly
memmapped.
- Return type:
ArrayLike
- load_frame(image_id)[source]¶
TODO: FINISHME or rename to lazy frame?
Returns a frame object that lazy loads on slice
- prepare(gids=None, workers=0, use_stamp=True)[source]¶
Precompute the cached frame conversions
- Parameters:
gids (List[int] | None) – specific image ids to prepare. If None prepare all images.
workers (int, default=0) – number of parallel threads for this io-bound task
Example
>>> from ndsampler.abstract_frames import * >>> workdir = ub.Path.appdir('ndsampler/tests/test_cog_precomp').ensuredir() >>> print('workdir = {!r}'.format(workdir)) >>> ub.delete(workdir) >>> ub.ensuredir(workdir) >>> self = SimpleFrames.demo(backend='npy', workdir=workdir) >>> print('self = {!r}'.format(self)) >>> print('self.cache_dpath = {!r}'.format(self.cache_dpath)) >>> #_ = ub.cmd('tree ' + workdir, verbose=3) >>> self.prepare() >>> self.prepare() >>> #_ = ub.cmd('tree ' + workdir, verbose=3) >>> _ = ub.cmd('ls ' + self.cache_dpath, verbose=3)
Example
>>> from ndsampler.abstract_frames import * >>> import ndsampler >>> workdir = ub.Path.appdir('ndsampler/tests/test_cog_precomp2') >>> workdir.delete() >>> # TEST NPY >>> # >>> sampler = ndsampler.CocoSampler.demo(workdir=workdir, backend='npy') >>> self = sampler.frames >>> ub.delete(self.cache_dpath) # reset >>> self.prepare() # serial, miss >>> self.prepare() # serial, hit >>> ub.delete(self.cache_dpath) # reset >>> self.prepare(workers=3) # parallel, miss >>> self.prepare(workers=3) # parallel, hit >>> # >>> ## TEST COG >>> # xdoctest: +REQUIRES(module:osgeo) >>> sampler = ndsampler.CocoSampler.demo(workdir=workdir, backend='cog') >>> self = sampler.frames >>> ub.delete(self.cache_dpath) # reset >>> self.prepare() # serial, miss >>> self.prepare() # serial, hit >>> ub.delete(self.cache_dpath) # reset >>> self.prepare(workers=3) # parallel, miss >>> self.prepare(workers=3) # parallel, hit
- class ndsampler.HashIdentifiable(**kwargs)[source]¶
Bases:
objectA class is hash-identifiable if its invariants can be tied to a specific list of hashable dependencies.
- The inheriting class must either:
implement _depends
implement _make_hashid
define _hashid
Example
- class Base:
- def __init__(self):
# commenting the next line removes cooperative inheritence super().__init__() self.base = 1
- class Derived(Base, HashIdentifiable):
- def __init__(self):
super().__init__() self.defived = 1
self = Derived() dir(self)
- property hashid¶
- exception ndsampler.MissingNegativePool[source]¶
Bases:
AssertionError
- class ndsampler.SimpleFrames(id_to_path, workdir=None, backend=None)[source]¶
Bases:
FramesBasic concrete implementation of frames objects for images where there is a strict one-file-to-one-image mapping (i.e. no auxiliary images).
- Parameters:
id_to_path (Dict) – mapping from image-id to image path
Example
>>> from ndsampler.abstract_frames import * >>> self = SimpleFrames.demo(backend='npy') >>> pathinfo = self._build_pathinfo(1) >>> print('pathinfo = {}'.format(ub.urepr(pathinfo, nl=3)))
>>> assert self.load_image(1).shape == (512, 512, 3) >>> assert self.load_region(1, (slice(-20), slice(-10))).shape == (492, 502, 3)
- property image_ids¶
- ndsampler.select_positive_regions(targets, window_dims=(300, 300), thresh=0.0, rng=None, verbose=0)[source]¶
Reduce positive example redundency by selecting disparate positive samples
Example
>>> from ndsampler.coco_regions import * >>> import kwcoco >>> dset = kwcoco.CocoDataset.demo('shapes8') >>> targets = tabular_coco_targets(dset) >>> window_dims = (300, 300) >>> selected = select_positive_regions(targets, window_dims) >>> print(len(selected)) >>> print(len(dset.anns))